Antibody data
- Antibody Data
- Antigen structure
- References [25]
- Comments [0]
- Validations
- Western blot [2]
- Other assay [22]
Submit
Validation data
Reference
Comment
Report error
- Product number
- 14-9858-80 - Provider product page

- Provider
- Invitrogen Antibodies
- Product name
- IRF4 Monoclonal Antibody (3E4), eBioscience™
- Antibody type
- Monoclonal
- Antigen
- Other
- Description
- Description: The monoclonal antibody 3E4 reacts with human and mouse interferon regulatory factor 4 (IRF4). IRF4 is a 52 kDa transcription factor with roles in B cell, T cell and macrophage function. In B cells, IRF4 is highly expressed in mature plasma cells and plays a crucial role in their differentiation. IRF4 has been shown to interact with PU.1 and control the transcription of many B cell-specific genes including Prdm1, which encodes Blimp1. In T cells, IRF4 has been implicated in regulatory T (Treg), T helper (Th)2, Th9 and Th17 cell development and function. This transcription factor is upregulated upon T cell activation and is expressed in mature T cells. Studies have shown that IRF4 directly induces Blimp1 expression in Tregs, leading to IL-10 expression. IRF4 has also been demonstrated to be involved in macrophage polarization and regulation. Lastly, in addition to its roles in normal immune function, IRF4 expression has been reported to be upregulated in many blood-related cancers. Applications Reported: This 3E4 antibody has been reported for use in western blotting. Applications Tested: This 3E4 antibody has been tested by western blot of reduced cell lysate prepared from splenocytes cultured for 3 days in the presence of plate-bound Anti-Mouse CD3 and soluble Anti-Mouse CD28. This antibody can be used at less than or equal to 5 µg/mL. It is recommended that the antibody be carefully titrated for optimal performance in the assay of interest. Purity: Greater than 90%, as determined by SDS-PAGE. Aggregation: Less than 10%, as determined by HPLC. Filtration: 0.2 µm post-manufacturing filtered.
- Reactivity
- Human, Mouse
- Host
- Rat
- Isotype
- IgG
- Antibody clone number
- 3E4
- Vial size
- 25 µg
- Concentration
- 0.5 mg/mL
- Storage
- 4° C
Submitted references B cells promote CD8 T cell primary and memory responses to subunit vaccines.
Mitochondrial Oxidative Phosphorylation Regulates the Fate Decision between Pathogenic Th17 and Regulatory T Cells.
Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR.
Pro-Survival Lipid Sphingosine-1-Phosphate Metabolically Programs T Cells to Limit Anti-tumor Activity.
CD4(+) T Cell Fate Decisions Are Stochastic, Precede Cell Division, Depend on GITR Co-Stimulation, and Are Associated With Uropodium Development.
Blimp-1 Functions as a Molecular Switch to Prevent Inflammatory Activity in Foxp3(+)RORγt(+) Regulatory T Cells.
ROCK2, but not ROCK1 interacts with phosphorylated STAT3 and co-occupies TH17/TFH gene promoters in TH17-activated human T cells.
Ubiquitin specific peptidase 4 stabilizes interferon regulatory factor protein and promotes its function to facilitate interleukin-4 expression in T helper type 2 cells.
RANKL-mediated harmonious dialogue between fetus and mother guarantees smooth gestation by inducing decidual M2 macrophage polarization.
Batf3 selectively determines acquisition of CD8(+) dendritic cell phenotype and function.
Affinity and dose of TCR engagement yield proportional enhancer and gene activity in CD4+ T cells.
STAT3/5-Dependent IL9 Overexpression Contributes to Neoplastic Cell Survival in Mycosis Fungoides.
Group 2 innate lymphoid cells utilize the IRF4-IL-9 module to coordinate epithelial cell maintenance of lung homeostasis.
γ-Tocopherol supplementation of allergic female mice augments development of CD11c+CD11b+ dendritic cells in utero and allergic inflammation in neonates.
Co-potentiation of antigen recognition: A mechanism to boost weak T cell responses and provide immunotherapy in vivo.
Cell-intrinsic expression of TLR9 in autoreactive B cells constrains BCR/TLR7-dependent responses.
Beta-catenin signaling drives differentiation and proinflammatory function of IRF8-dependent dendritic cells.
Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice.
OX40 signaling favors the induction of T(H)9 cells and airway inflammation.
The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells.
The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection.
Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells.
IRF4: Immunity. Malignancy! Therapy?
Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses.
A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells.
Klarquist J, Cross EW, Thompson SB, Willett B, Aldridge DL, Caffrey-Carr AK, Xu Z, Hunter CA, Getahun A, Kedl RM
Cell reports 2021 Aug 24;36(8):109591
Cell reports 2021 Aug 24;36(8):109591
Mitochondrial Oxidative Phosphorylation Regulates the Fate Decision between Pathogenic Th17 and Regulatory T Cells.
Shin B, Benavides GA, Geng J, Koralov SB, Hu H, Darley-Usmar VM, Harrington LE
Cell reports 2020 Feb 11;30(6):1898-1909.e4
Cell reports 2020 Feb 11;30(6):1898-1909.e4
Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR.
Zhu H, Bhatt B, Sivaprakasam S, Cai Y, Liu S, Kodeboyina SK, Patel N, Savage NM, Sharma A, Kaufman RJ, Li H, Singh N
Nature communications 2019 Mar 6;10(1):1084
Nature communications 2019 Mar 6;10(1):1084
Pro-Survival Lipid Sphingosine-1-Phosphate Metabolically Programs T Cells to Limit Anti-tumor Activity.
Chakraborty P, Vaena SG, Thyagarajan K, Chatterjee S, Al-Khami A, Selvam SP, Nguyen H, Kang I, Wyatt MW, Baliga U, Hedley Z, Ngang RN, Guo B, Beeson GC, Husain S, Paulos CM, Beeson CC, Zilliox MJ, Hill EG, Mehrotra M, Yu XZ, Ogretmen B, Mehrotra S
Cell reports 2019 Aug 13;28(7):1879-1893.e7
Cell reports 2019 Aug 13;28(7):1879-1893.e7
CD4(+) T Cell Fate Decisions Are Stochastic, Precede Cell Division, Depend on GITR Co-Stimulation, and Are Associated With Uropodium Development.
Cobbold SP, Adams E, Howie D, Waldmann H
Frontiers in immunology 2018;9:1381
Frontiers in immunology 2018;9:1381
Blimp-1 Functions as a Molecular Switch to Prevent Inflammatory Activity in Foxp3(+)RORγt(+) Regulatory T Cells.
Ogawa C, Bankoti R, Nguyen T, Hassanzadeh-Kiabi N, Nadeau S, Porritt RA, Couse M, Fan X, Dhall D, Eberl G, Ohnmacht C, Martins GA
Cell reports 2018 Oct 2;25(1):19-28.e5
Cell reports 2018 Oct 2;25(1):19-28.e5
ROCK2, but not ROCK1 interacts with phosphorylated STAT3 and co-occupies TH17/TFH gene promoters in TH17-activated human T cells.
Chen W, Nyuydzefe MS, Weiss JM, Zhang J, Waksal SD, Zanin-Zhorov A
Scientific reports 2018 Nov 9;8(1):16636
Scientific reports 2018 Nov 9;8(1):16636
Ubiquitin specific peptidase 4 stabilizes interferon regulatory factor protein and promotes its function to facilitate interleukin-4 expression in T helper type 2 cells.
Guo Z, Xu P, Ge S, Zhang C, Zheng X, Xu J, Liu Z, Li B, Ge S
International journal of molecular medicine 2017 Oct;40(4):979-986
International journal of molecular medicine 2017 Oct;40(4):979-986
RANKL-mediated harmonious dialogue between fetus and mother guarantees smooth gestation by inducing decidual M2 macrophage polarization.
Meng YH, Zhou WJ, Jin LP, Liu LB, Chang KK, Mei J, Li H, Wang J, Li DJ, Li MQ
Cell death & disease 2017 Oct 12;8(10):e3105
Cell death & disease 2017 Oct 12;8(10):e3105
Batf3 selectively determines acquisition of CD8(+) dendritic cell phenotype and function.
Chandra J, Kuo PT, Hahn AM, Belz GT, Frazer IH
Immunology and cell biology 2017 Feb;95(2):215-223
Immunology and cell biology 2017 Feb;95(2):215-223
Affinity and dose of TCR engagement yield proportional enhancer and gene activity in CD4+ T cells.
Allison KA, Sajti E, Collier JG, Gosselin D, Troutman TD, Stone EL, Hedrick SM, Glass CK
eLife 2016 Jul 4;5
eLife 2016 Jul 4;5
STAT3/5-Dependent IL9 Overexpression Contributes to Neoplastic Cell Survival in Mycosis Fungoides.
Vieyra-Garcia PA, Wei T, Naym DG, Fredholm S, Fink-Puches R, Cerroni L, Odum N, O'Malley JT, Gniadecki R, Wolf P
Clinical cancer research : an official journal of the American Association for Cancer Research 2016 Jul 1;22(13):3328-39
Clinical cancer research : an official journal of the American Association for Cancer Research 2016 Jul 1;22(13):3328-39
Group 2 innate lymphoid cells utilize the IRF4-IL-9 module to coordinate epithelial cell maintenance of lung homeostasis.
Mohapatra A, Van Dyken SJ, Schneider C, Nussbaum JC, Liang HE, Locksley RM
Mucosal immunology 2016 Jan;9(1):275-86
Mucosal immunology 2016 Jan;9(1):275-86
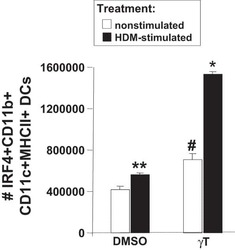
γ-Tocopherol supplementation of allergic female mice augments development of CD11c+CD11b+ dendritic cells in utero and allergic inflammation in neonates.
Abdala-Valencia H, Soveg F, Cook-Mills JM
American journal of physiology. Lung cellular and molecular physiology 2016 Apr 15;310(8):L759-71
American journal of physiology. Lung cellular and molecular physiology 2016 Apr 15;310(8):L759-71
Co-potentiation of antigen recognition: A mechanism to boost weak T cell responses and provide immunotherapy in vivo.
Hoffmann MM, Molina-Mendiola C, Nelson AD, Parks CA, Reyes EE, Hansen MJ, Rajagopalan G, Pease LR, Schrum AG, Gil D
Science advances 2015 Oct;1(9):e1500415
Science advances 2015 Oct;1(9):e1500415
Cell-intrinsic expression of TLR9 in autoreactive B cells constrains BCR/TLR7-dependent responses.
Nündel K, Green NM, Shaffer AL, Moody KL, Busto P, Eilat D, Miyake K, Oropallo MA, Cancro MP, Marshak-Rothstein A
Journal of immunology (Baltimore, Md. : 1950) 2015 Mar 15;194(6):2504-12
Journal of immunology (Baltimore, Md. : 1950) 2015 Mar 15;194(6):2504-12
Beta-catenin signaling drives differentiation and proinflammatory function of IRF8-dependent dendritic cells.
Cohen SB, Smith NL, McDougal C, Pepper M, Shah S, Yap GS, Acha-Orbea H, Jiang A, Clausen BE, Rudd BD, Denkers EY
Journal of immunology (Baltimore, Md. : 1950) 2015 Jan 1;194(1):210-22
Journal of immunology (Baltimore, Md. : 1950) 2015 Jan 1;194(1):210-22
Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice.
Watchmaker PB, Lahl K, Lee M, Baumjohann D, Morton J, Kim SJ, Zeng R, Dent A, Ansel KM, Diamond B, Hadeiba H, Butcher EC
Nature immunology 2014 Jan;15(1):98-108
Nature immunology 2014 Jan;15(1):98-108
OX40 signaling favors the induction of T(H)9 cells and airway inflammation.
Xiao X, Balasubramanian S, Liu W, Chu X, Wang H, Taparowsky EJ, Fu YX, Choi Y, Walsh MC, Li XC
Nature immunology 2012 Oct;13(10):981-90
Nature immunology 2012 Oct;13(10):981-90
The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells.
Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, Kallies A
Nature immunology 2011 Apr;12(4):304-11
Nature immunology 2011 Apr;12(4):304-11
The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection.
Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S
Nature immunology 2010 Oct;11(10):936-44
Nature immunology 2010 Oct;11(10):936-44
Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells.
Staudt V, Bothur E, Klein M, Lingnau K, Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Ulges A, Taube C, Dehzad N, Becker M, Stassen M, Steinborn A, Lohoff M, Schild H, Schmitt E, Bopp T
Immunity 2010 Aug 27;33(2):192-202
Immunity 2010 Aug 27;33(2):192-202
IRF4: Immunity. Malignancy! Therapy?
Shaffer AL, Emre NC, Romesser PB, Staudt LM
Clinical cancer research : an official journal of the American Association for Cancer Research 2009 May 1;15(9):2954-61
Clinical cancer research : an official journal of the American Association for Cancer Research 2009 May 1;15(9):2954-61
Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses.
Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY
Nature 2009 Mar 19;458(7236):351-6
Nature 2009 Mar 19;458(7236):351-6
A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells.
Falini B, Fizzotti M, Pucciarini A, Bigerna B, Marafioti T, Gambacorta M, Pacini R, Alunni C, Natali-Tanci L, Ugolini B, Sebastiani C, Cattoretti G, Pileri S, Dalla-Favera R, Stein H
Blood 2000 Mar 15;95(6):2084-92
Blood 2000 Mar 15;95(6):2084-92
No comments: Submit comment
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
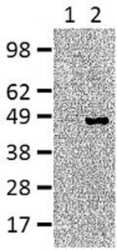
- Experimental details
- Immunoblotting of reduced cell lysate prepared from mouse splenocytes with 5 µg/mL of Anti-Human/Mouse IRF4 antibody.Splenocytes were cultured for 3 days in either complete media alone (Lane 1) or polarized under Th2 conditions (Lane 2).Bands were visualized using Anti-Mouse IgG HRP.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Western blot was performed using Anti-IRF4 Monoclonal Antibody (3E4) (Product # 14-9858-82) and a 51kDa band corresponding to IRF4 was observed in IM9 and was absent in MOLT-4, Jurkat and U-937 as reported. Whole cell extracts (30 µg lysate) of IM9 (Lane 1), MOLT-4 (Lane 2), Jurkat (Lane 3) and U-937 (Lane 4) were electrophoresed using Novex® NuPAGE® 4-12 % Bis-Tris gel (Product # NP0322BOX). Resolved proteins were then transferred onto a nitrocellulose membrane (Product # IB23001) by iBlot® 2 Dry Blotting System (Product # IB21001). The blot was probed with the primary antibody (5ug/ml) and detected by chemiluminescence F(ab)2-Rabbit anti-Rat IgG (H+L) Secondary Antibody, HRP (Product # PA1-29927; 1:4000 dilution) using the iBright FL 1000 (Product # A32752). Chemiluminescent detection was performed using Novex® ECL Chemiluminescent Substrate Reagent Kit (Product # WP20005).
Supportive validation
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
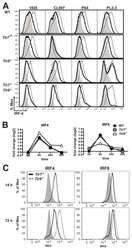
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
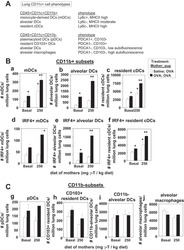
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
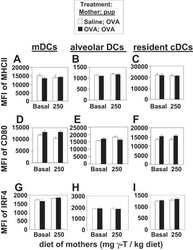
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
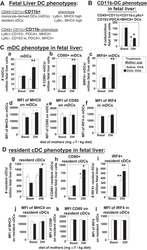
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
- NULL
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
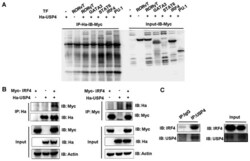
- Figure 1 Interaction between ubiquitin specific peptidase 4 (USP4) and interferon regulatory factor 4 (IRF4). (A) IRF4 was co-immunoprecipitated by the anti-Ha antibody when Ha-USP4 and Myc-IRF4 were coexpressed. (B) IRF4 was co-immunoprecipitated by the anti-Ha antibody when Ha-USP4 and Myc-IRF4 were coexpressed, and USP4 was co-immunoprecipitated by the anti-Myc antibody when Ha-USP4 and Myc-IRF4 were coexpressed. (C) IRF4 was co-immunoprecipitated by the anti-USP4 antibody, indicating that USP4 interacts with IRF4 in Th2 cells. Flag-IRF4 and Ha-USP4 were co-transfected into 293T cells. After 48 h, the cells were retrieved, and 1 u g of anti-Ha or anti-Flag monoclonal antibody precipitate was added and immunoblotting was conducted in A and B. Th2 cells were amplified to 1x10 7 , and co-immunoprecipitated with 1 u g of normal IgG or anti-USP4, then immunoblotting assay was conducted in C.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
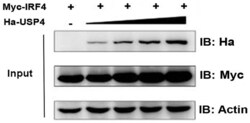
- Figure 2 Effect of ubiquitin specific peptidase 4 (USP4) on stabilization of interferon regulatory factor 4 (IRF4). IRF4 protein expression level was influenced by USP4 in a dose-dependent manner. Equivalent dose of Myc-IRF4 and different doses of Ha-USP4 were cotransfected into 293T cells. After 48 h, the cells were retrieved and the expression levels of USP4 and IRF4 were detected by immunoblotting.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
- Figure 4 Ubiquitin specific peptidase 4 (USP4) and interferon regulatory factor 4 (IRF4) synergize with nuclear factor of activated T cell-2 (NFATc2) to specifically enhance NFAT-mediated activation of the interleukin-4 (IL-4) promoter. (A) Cotransfection with USP4 and IRF4 had no significant effects on the luciferase activity of IL-4-luc. (B and C) Cotransfection with USP4, IRF4 and NFATc2 markedly increased the luciferase activity of IL-4-luc compared with that of the controls. * p
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
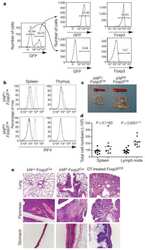
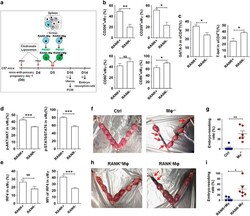
- Figure 5 Adoptive transfer of RANK + M phi relieves mouse embryo absorption induced by M phi depletion. ( a ) RANK + and RANK - M phi s were isolated from mouse spleen, labeled with PKH-67, and then transferred to M phi -depleted pregnant mice at day 5 of gestation. The uterus was then collected and analyzed by FCM at day 10, and the embryo resorption ratio was observed at day 14. ( b ) FCM analysis of CD206, CD209, CD80 and CD86 in PKH-67-uM phi with PKH-67-RANK + M phi and PKH-67-RANK - M phi transfer at day 10 ( n =5 mice per group). (Student''s t -test). ( c ) FCM analysis of GATA-3 and T-bet in PKH-67-uM phi with PKH-67-RANK + M phi and PKH-67-RANK - M phi transfer at day 10 ( n =5 mice per group); (Student''s t -test). ( d ) FCM analysis of the phosphorylation level of Akt and STAT6 in uM phi cells with PKH-67-RANK + M phi and PKH-67-RANK - M phi transfer at day 10 ( n =5 mice per group); (Student''s t -test). ( e ) FCM analysis of the percentage and median fluorescence intensity (MFI) of IRF4 in uM phi cells with PKH-67-RANK + M phi and PKH-67-RANK - M phi transfer at day 10 ( n =5 mice per group); (Student''s t -test). ( f and g ) The embryo absorption rate in ctrl pregnant C57BL/6 mice and pregnant C57BL/6 mice with M phi depletion ( n =6 mice per group) were counted on day 14 of gestation (adjusted t -test). ( h and i ) The embryo absorption rate in pregnant C57BL/6 mice with M phi depletion ( n =6 mice per group) was counted on day 14 of gestation, after adoptive t
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
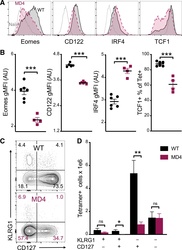
- Figure 4. B cells promote CD8 T cell memory programming following subunit vaccination (A) Representative histograms for Eomes, CD122, IRF4, and TCF1 staining of tetramer+ cells from WT (black) or MD4 (maroon) mice 7 days after combined-adjuvant vaccination or for CD44- naive control CD8 T cells (gray). (B) The gMFI of tetramer+ cells for Eomes, CD122, and IRF4, where the x axis intersects the y axis at the average gMFI value for CD44- naive controls, and the percentage of tetramer+ cells that stained positive for TCF1. (C and D) Representative KLRG1 versus CD127 staining (C) and numbers of tetramer+ splenocytes in the respective quadrants (D), 7 days after immunization. Data shown are means +- SEM; n : mice per group, representative of 3 experiments. Significance was defined by two-tailed, unpaired Student's t tests, where *p < 0.05, **p < 0.01, and ***p < 0.001.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
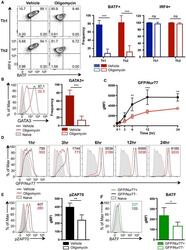
- Figure 4. ATP-Linked OXPHOS Controls BATF Induction by Regulating TCR Signaling (A and B) Naive CD4 T cells were differentiated under Th1 or Th2 polarizing conditions in the presence of vehicle or oligomycin for 24 h, and expression of (A) BATF, IRF4, and (B) GATA3 was determined by flow cytometry (4 to 5 independent experiments). (C and D) Naive CD4 T cells from Nur77/GFP spleens were activated under Th17 conditions in the presence of vehicle or oligomycin for indicated period of time. (C) Graph shows the expression of Nur77/GFP. (D) Representative plots are gated on live CD4 T cells (representative of 3 independent experiments). (E) Naive CD4 T cells were activated under Th17 conditions with vehicle or oligomycin. Representative histogram overlay shows the phosphorylation of ZAP70 at 24 h (representative of 5 independent experiments). (F) Naive CD4 T cells from Nur77/GFP reporter spleens were stimulated with Th17 cytokines for 6 h. BATF expression in GFP + and GFP - cells were determined by flow cytometry (3 independent experiments). Graphs show the average +- SD; (A and C) two-way ANOVA; (B, E and F) unpaired t test *p < 0.05, **p < 0.01, ***p < 0.001.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
- Experimental details
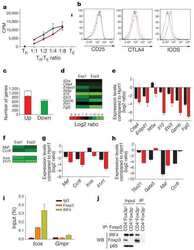
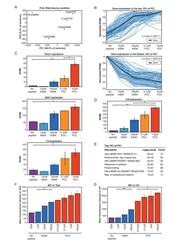
- Figure 2. RNA-Sequencing reveals graded expression of activation signature genes. ( A ) Principal component analysis (PCA) of the approximately 3,200 genes that changed between any two samples reveals that the primary axis of variation (PC1, shown along the x-axis) orders the five conditions by increasing TCR signal strength: No Peptide; low-dose, low-affinity (10 muM K99A); high-dose, low-affinity (100 muM K99A); low-dose, high-affinity (0.1 muM PCC); and high-dose, high-affinity (10 muM PCC). ( B ) After ordering the ~3200 genes used for PCA by their contribution to PC1, we extracted the top 10%--that is, the ~320 genes contributing most positively to a sample's PC1 value--and the bottom 10%--that is, the ~320 genes contributing most negatively to a sample's PC1 value. Each group displays a clear trend, with the top 10% increasing in expression as signal strength increases, and the bottom 10% decreasing in expression. Each blue line represents a gene, with reads per kilobase per million (RPKM) normalized from 0 to 1 across the five conditions. Significance was determined using permutation testing, where the mean difference between genes in the No Peptide sample as compared to 10 muM PCC was normally distributed over randomly generated groups of genes. This normal distribution was compared to the top 10% and bottom 10% genes to generate a p-value. ( C ) Genes in the top 10% of PC1, termed activation signature genes, include many genes previously identified as impor
- Submitted by
- Invitrogen Antibodies (provider)
- Main image

- Experimental details
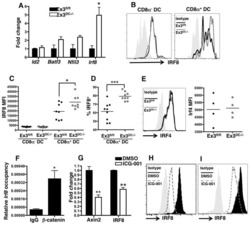
- Figure 4 IRF4-expressing CD8+ DCs in Batf3-/- mice are able to process antigen but lose capacity to take up dying cells (a) Splenocytes of Batf3-/- and control mice were incubated with DQ-OVA for 3 hours and subsequently analysed by flow cytometry for DQ-OVA processing. CD8+ and CD11b+ dendritic cell subsets were gated as shown in Figure 2 . Shown are representative histograms of green fluorescence (DQ-OVA) of treated versus untreated CD8+ and CD11b+ DCs in Batf3-/- and control splenocytes. The percentage and MFI of DQ-OVA+ CD8+ or CD11b+ DCs as well as of untreated samples (-) was compared. Each point represents a sample of one animal with indication of mean +/- SD of whole group (n=4). Shown is one of two independent experiments. (b) Splenocytes of Batf3-/- and control mice were incubated with DiI-labelled liposomes for 90 min and subsequently analysed by flow cytometry for liposome uptake. Shown are representative histograms of red fluorescence (DiI+) of CD8+ and CD11b+ DCs in Batf3-/- and control splenocytes incubated at 37degC or on ice. The percentage of DiI+ cells of CD8+ or CD11b+ DCs was compared. Each point represents a sample of one animal with indication of mean+/- SD of the whole group (n=5). (c-d) CellTrace Violet labelled, UV irradiated dying cells were injected into Batf3-/- and control mice. 3 hours later, splenic CD11b+ and CD8+ DCs were analysed for endocytosis of dying cells. (c) Shown are flow cytometry plots that were pre-gated on DCs as shown in Supplem
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
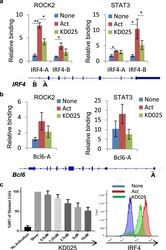
- Experimental details
- Figure 3 Targeted ROCK2 inhibition decreased the binding of ROCK2 to the Irf4 and Bcl6 promoters at Th17-skewing conditions. Human peripheral blood CD4 + T cells were stimulated under Th17-skewing conditions for 48 hours in the absence or presence of KD025, chromatin was purified and proceeded to ChIP-qPCR analyses with anti-ROCK2 or anti-STAT3 antibodies ( a , b ), or Cells were stained with antibodies to IRF4 and analyzed by Flow Cytometry ( c , d ). ( c ) IRF4 expression measured by flow cytometry. Left, Mean Fluorescence Index (MFI) for IRF4 is plotted as percentage of MFI for skewed cells (as 100%). Right, Histogram of the flow cytometry analysis. The average of four different experiments is shown. *p < 0.05; **p < 0.01.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
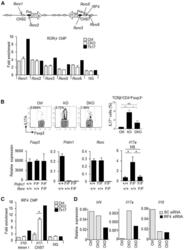
- Experimental details
- Figure 3. Increased Binding of IRF4 at the Il17 Locus and IRF4 and RORgammat Requirement for IL-17A Production in Blimp-1 -/- Foxp3 + Treg (A) Representation of the Il17 locus showing RORgammat and IRF4 consensus binding sites (top) and RORgt ChIP (bottom) in Blimp-1 +/+ and Blimp-1 -/- CD4 + Foxp3 GFP+ Treg and wild-type pTh17 cells. (B) IL-17A expression in TCRbeta + CD4 + Foxp3 + Treg (MLNs) from Prdm1 +/+ Rorc +/+ Foxp3 CREYFP+ (Ctrl), Prdm1 F/F Rorc +/+ Foxp 3CREYFP+ (knockout [KO]), and Prdm1 F/F Rorc F/F Foxp3 CREYFP+ (double knockout [DKO]) mice and expression of Foxp3 , Prdm1 , Rorc , and Il17a mRNA (bottom graphs) in sorted and in vitro -stimulated (phorbol myristate acetate [PMA] and ionomycin for 4 hr) CD4 + Foxp3 CREYFP+ cells. (C) IRF4 ChIP at the Il17 CNS7 and Il10 Intron I regions in Blimp1 +/+ and Blimp1 -/- CD4 + Foxp3 GFP+ Treg and wild-type pTh17 cells. (D) Irf4 , Il17a , and Il10 mRNA expression following Irf4 siRNA knockdown (SC, scrambled control). Data shown are representative of at least two independent experiments. Error bars show SEM (n >= 3 per group). *p < 0.05 and **p < 0.01, one-way ANOVA.
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
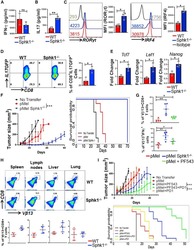
- Experimental details
- Figure 2. SphK1 -/- T Cells Secrete Both IFN-gamma and IL-17 upon TCR Stimulation (A-C) pMel and pMel-SphK -/- T cells activated for 3 days with cognate antigen gp100 were antigen-restimulated overnight, and the supernatant was used for ELISA-based determination of secreted cytokine: (A) IFN-gamma, (B) IL-17, (C) RORgammat and IRF4 levels. (D) Splenic T cells from IL-17-GFP reporter or SphK1 -/- -IL-17-GFP mice were cultured in Th17 polarizing condition for 3 days, restimulated with anti-CD3 (2 ug/mL) and anti-CD28 antibody (5 ug/mL) for 4 h, and the percentage of IL-17-GFP-positive cells were determined by flow cytometry. (E) Three-day TCR-activated WT and SphK1 -/- T cells were analyzed for expression of stemness-associated genes using mRNA. Data are representative of three independent experiments in (A)-(E). (F) C57BL/6 mice (n = 10 mice/group) were inoculated subcutaneously (s.c.) with 0.3 x 10 6 B16-F10 melanoma cells for 9 days, after which mice were either kept untreated or adoptively transferred, either with 1 x 10 6 pMel or pMel-SphK1 -/- T cells. Tumor growth was measured using digital calipers twice weekly. Left panel: estimated growth curves for each experimental condition. Right panel: survival curve (p value pMel versus pMel SphK1 -/- is 1e-05). (G) Upper panel: C57BL/6 mice (n = 5 mice/group) were inoculated (s.c.) with 0.25 x 10 6 B16-F10 melanoma cells for 14 days, then 1 x 10 6 3 days gp100-activated pMel T cells, pMel SphK1 -/- T cells, or pMel T cells trea
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
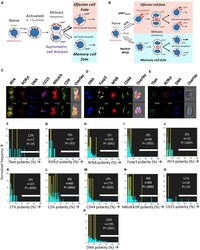
- Experimental details
- Figure 6 The CD4 + T cell fate choice associated with uropodium development does not depend on asymmetric cell divisions. (A) Depiction of a binary cell fate decision as a result of an asymmetric first cell division. The effector and memory cell fates result from a differential inheritance of mitochondria, CD4, and PI3k signaling between the two daughters. (B) A stochastic cell fate decision to develop uropodia during initial activation and before any cell division. The chance of any individual cell becoming either an effector cell, and developing uropodia, or a memory cell without uropodia, depends on the balance of a number of interacting signaling pathways (e.g., GITR, mTORC1, mTORC2, and NFkappaB) during its initial activation. Symmetrical cell divisions can then take place regardless of the cell fate decision taken. (C-O) CTV labeled naive CD4 + T cells from female A1RAG were stimulated with bone marrow derived dendritic cells + 100 nM Dby peptide with IL-2, TGFbeta, and ATRA for 48 h (when the average number of cell divisions was only 0.7). No mitotic inhibitors were used. Cultures were labeled "" in situ "" with (C,L) CD4-APC-Cy7 and CD25-BV605 or (D,M) CD44-APC-eflour780, plus live/dead aqua and Mitotracker DR (C,E,N) , and fixed at 37degC by the addition, without mixing or disturbing the cultures, of 40% formaldehyde to 10% v/v for 15 min. The medium and fixative were then carefully aspirated and fix/perm buffer added for 2 h at 37degC. Intracellular stains used were
- Submitted by
- Invitrogen Antibodies (provider)
- Main image
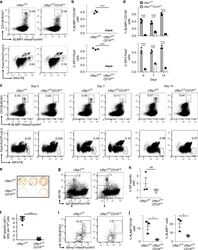
- Experimental details
- Fig. 2 Decreased number of plasma cells in Ufbp1 F/F CD19 cre mice. a Bone marrow cells from indicated mice were stained with BLIMP1, CD138, IRF4, and Pax5, and analyzed by flow cytometry. Shown is the BLIMP1 versus CD138 (upper panel), and IRF4 versus Pax5 staining (lower panel) b Enumeration of frequencies of BLIMP1 + CD138 + and IRF4 + Pax5 - cells in a ( n = 3 mice/genotype). c Ufbp1 F/F and Ufbp1 F/F CD19 cre mice immunized with 4-hydroxy-3-nitrophenylacetyl conjugated to keyhole limpet hemocyanin (NP-KLH) as in Fig. 1c were analyzed for plasma cells (BLIMP1 + CD138 + and IRF4 + Pax5 - ) in bone marrow by flow cytometry at the indicated time points after immunization. d Enumeration of the frequencies of plasma cells (BLIMP1 + CD138 + and IRF4 + Pax5 - ) in c ( n = 3 mice/genotype). e Indicated mice were immunized as in Fig. 1c . Twenty-one days later, bone marrow cells were analyzed for the presence of NP-specific IgG1 + ASCs by ELISpot. Shown is a photograph of representative ELISpot wells. f Number (mean +- SEM) of NP-specific IgG1 + ELISpots in e ( n = 6 mice/genotype). g Ufbp1 F/F and Ufbp1 F/F CD19 cre mice were immunized with NP-KLH in Sigma Adjuvant System and on day 10, 7-AAD - CD4 - CD8 - Gr-1 - CD11b - splenic cells were analyzed for IgD and NP staining by flow cytometry. h Enumeration of the frequency of IgD - NP + cells in g ( n = 3 mice/genotype). i BLIMP1 and CD138 expression by IgD - NP + cells in g was an
 Explore
Explore Validate
Validate Learn
Learn Western blot
Western blot